Technical Sharing | Breaking Through the Peanut Breeding Bottleneck, KASP Technology Aids in Precise Molecular Marker-Assisted Breeding
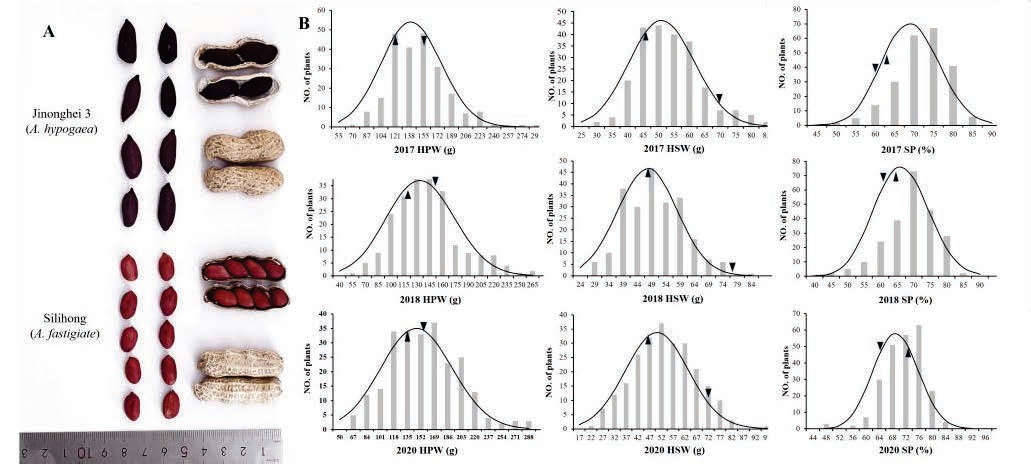
A recombinant inbred line (RIL) population was used, which originated from the cross between "Silihong" (A. hypogaea var. fastigiate) and "Jinonghei 3" (A. hypogaea var. hypogaea).
The U.S. mini-core germplasm collection was used for association analysis.
High-density genetic linkage map construction: A high-density genetic linkage map was constructed through resequencing of RILs, including 4499 bins covering 20 chromosomes with a total length of 1712.32 cM.
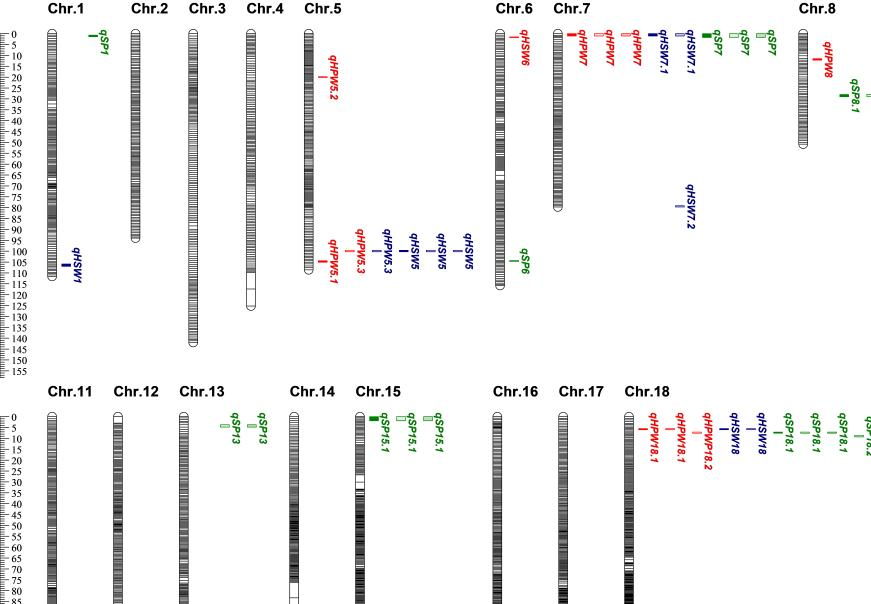
Quantitative Trait Loci (QTL) Analysis: 46 QTLs related to HPW, HSW, and SP were identified across three different environments. Some major QTLs, such as qHPW5.2, qSP7.1, and qSP18.2, showed high phenotypic variation explained rate (PVE).
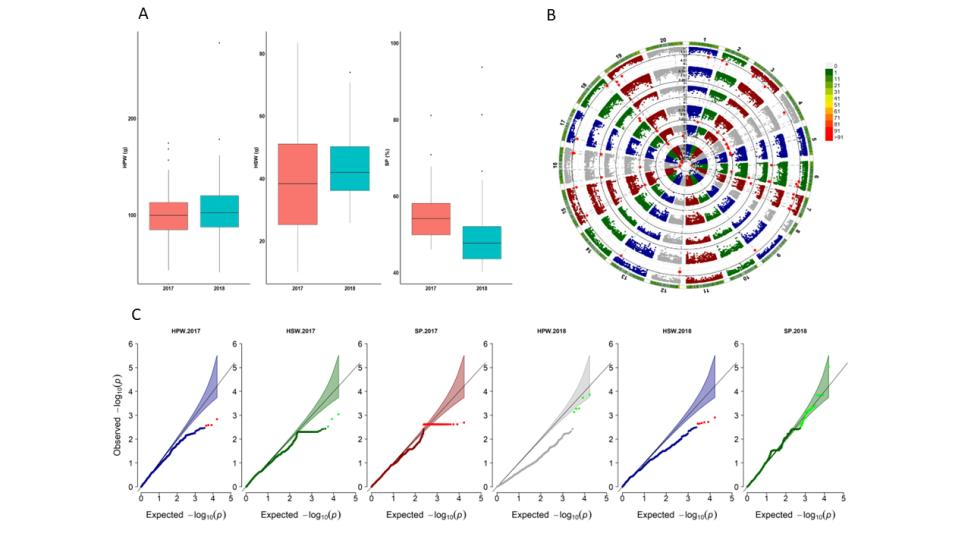
Genome-Wide Association Study (GWAS): GWAS was conducted using the U.S. mini-core germplasm collection to verify the accuracy of QTL positioning and identified 115 significantly associated single nucleotide polymorphisms (SNPs) with HPW, HSW, and SP in two environments.
KASP Marker Development and Validation: Three KASP markers were developed based on QTL regions and validated in peanut varieties and local varieties, showing significant association with HPW, HSW, and SP.
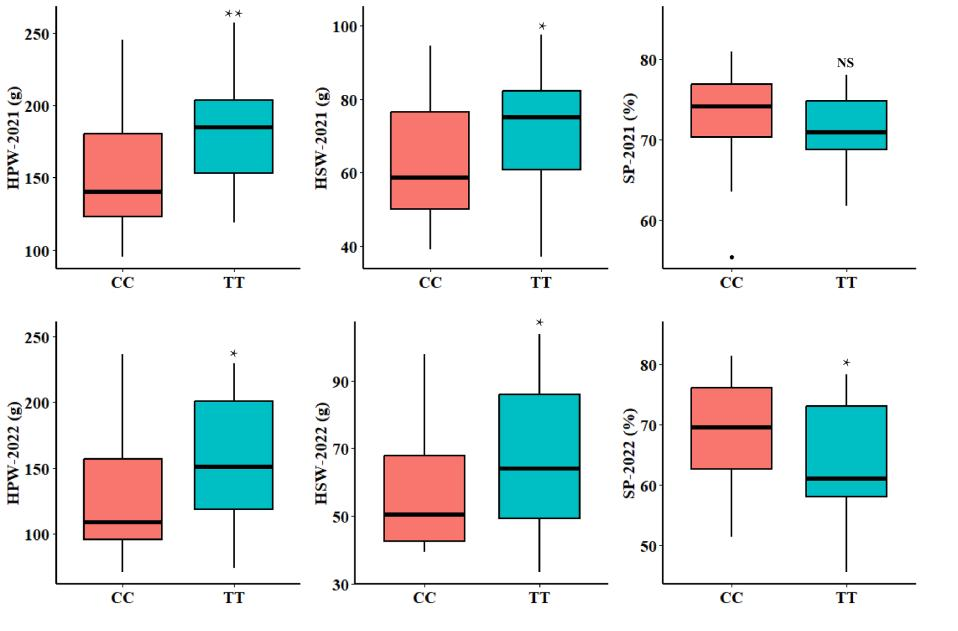
"The above figure demonstrates the impact of the KASP-SP7 marker on SP, HPW, and HSW in the peanut group. It includes statistical charts showing the effect of different alleles of KASP-SP7 on the target traits, with asterisks indicating the level of significance."
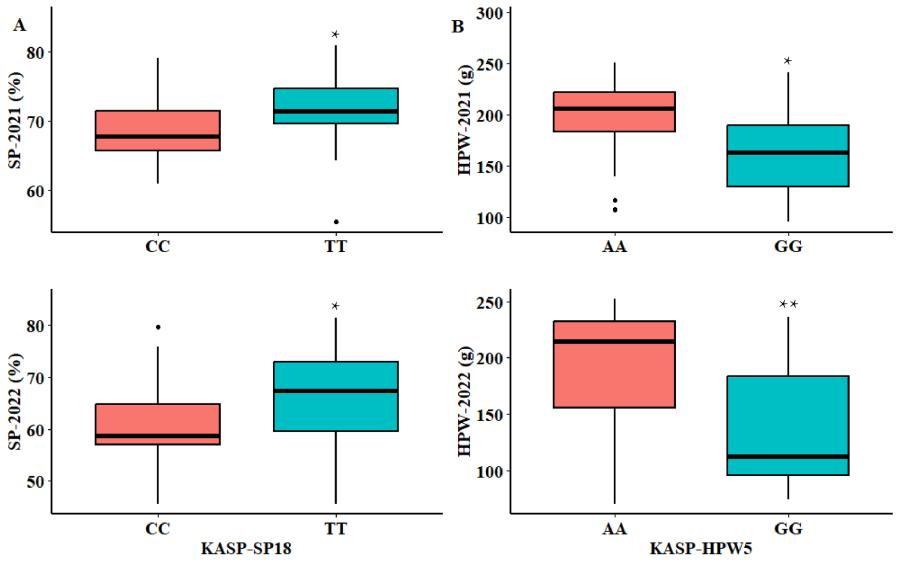
"The figure above shows the validation effects of the KASP-SP18 and KASP-HPW5 markers in the peanut group. It includes two sub-figures: A shows the impact of KASP-SP18 on SP, and B shows the impact of KASP-HPW5 on HPW, both using statistical charts and significance markers."
Multiple QTLs and SNPs related to HPW, HSW, and SP were identified, some of which were discovered for the first time.
GWAS validated the accuracy of QTL positioning and found some co-location of QTLs and SNPs.
The developed KASP markers provide a useful tool for peanut molecular-assisted breeding.
High Efficiency: KASP technology can significantly shorten the breeding cycle and improve breeding efficiency.
High Accuracy: SNP-based molecular marker technology ensures the accuracy of screening.
High Universality: KASP technology is suitable for molecular marker-assisted breeding in various crops.
Easy Operation: Simplifies the experimental operation process, making it easy to promote in different laboratories.
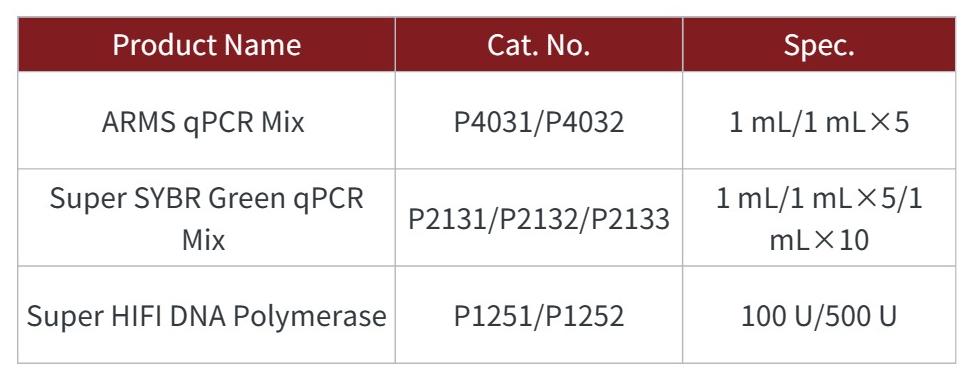
GDSBIO's Related Products: GDSBIO has developed several PCR Mix solutions for KASP technology, including hot-start Taq DNA polymerase with dual antibody modification, dNTPs, buffer, and other essential components for PCR amplification. The products have the advantages of easy use, superior performance, and low cost, and support trial applications.
KASP PCR Mix (click for details)
KASP PCR MixPlus (click for details)
 Tel: +86 20 31600213
Tel: +86 20 31600213  Sales EMail: order@gdsbio.com
Sales EMail: order@gdsbio.com